When a client is identified with cancer, among the most crucial actions is evaluation of the growth under a microscopic lense by pathologists to identify the cancer phase and to identify the growth. This info is main to comprehending medical diagnosis (i.e., most likely client results) and for identifying the most suitable treatment, such as going through surgical treatment alone versus surgical treatment plus chemotherapy. Establishing artificial intelligence (ML) tools in pathology to help with the tiny evaluation represents an engaging research study location with numerous prospective applications.
Previous research studies have actually revealed that ML can precisely determine and categorize growths in pathology images and can even anticipate client diagnosis utilizing understood pathology functions, such as the degree to which gland looks differ typical While these efforts concentrate on utilizing ML to find or measure recognized functions, alternative techniques use the prospective to determine unique functions. The discovery of brand-new functions might in turn even more enhance cancer prognostication and treatment choices for clients by drawing out info that isn’t yet thought about in present workflows.
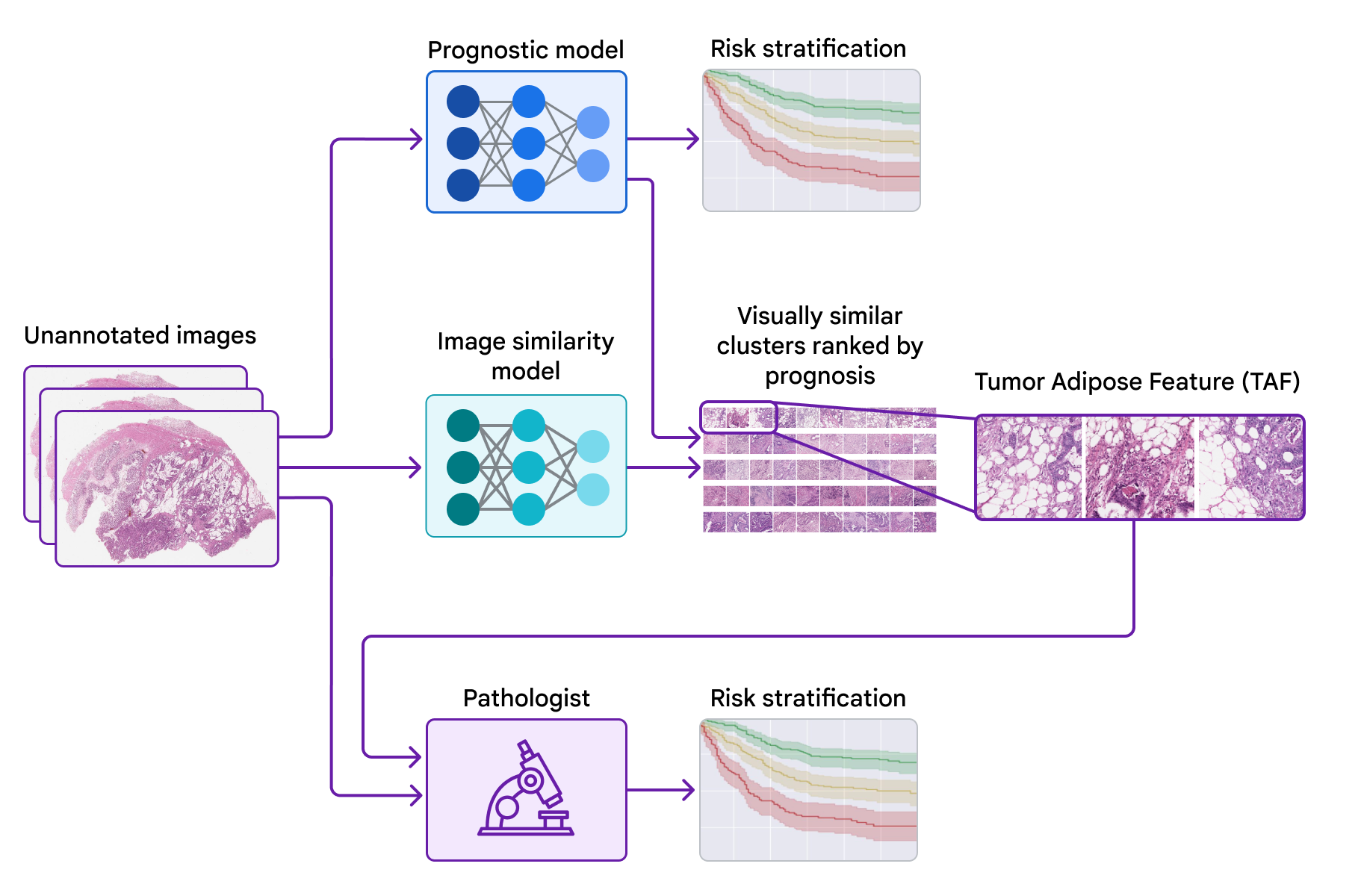
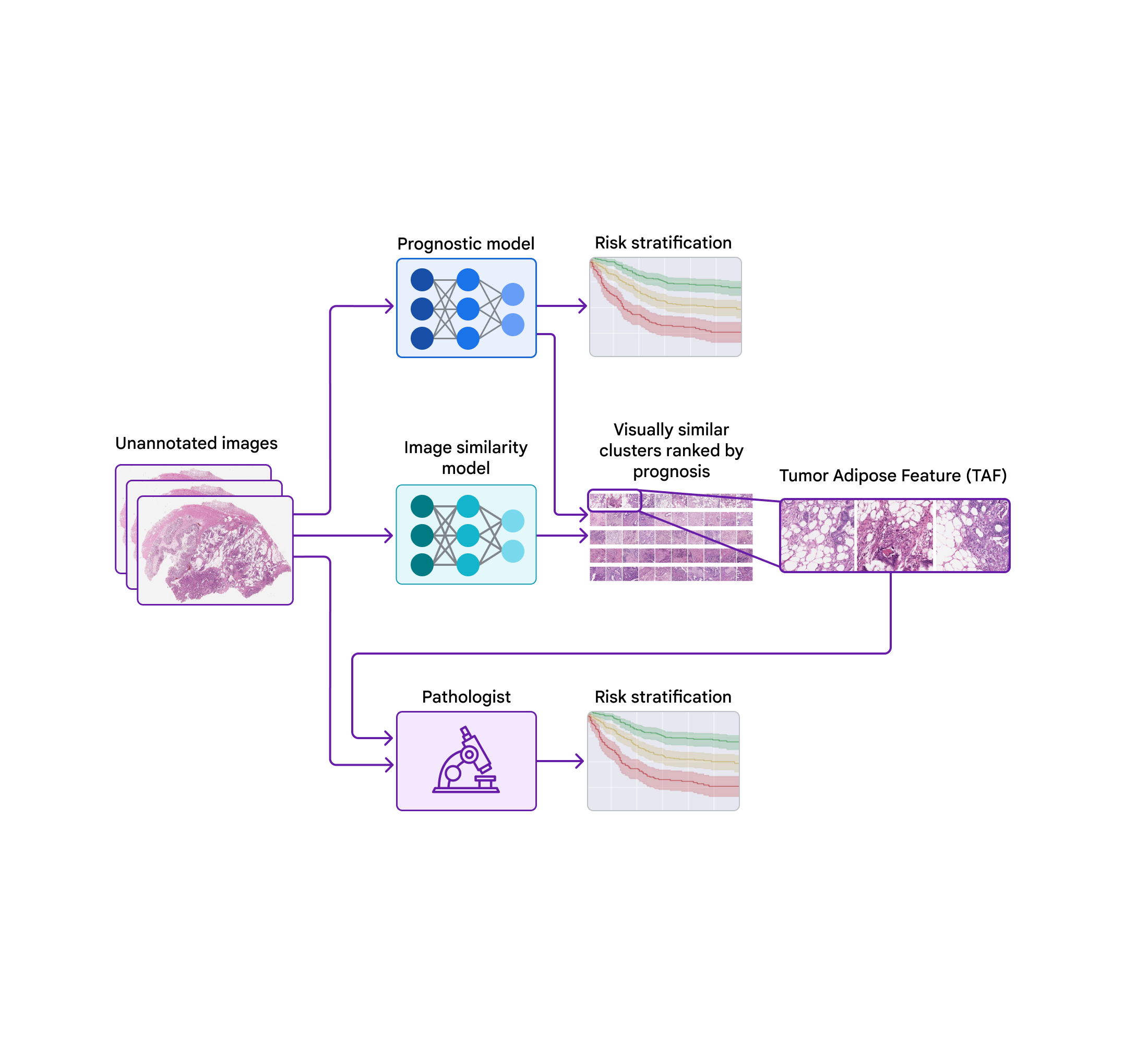
Today, we wish to share development we have actually made over the previous couple of years towards recognizing unique functions for colorectal cancer in partnership with groups at the Medical University of Graz in Austria and the University of Milano-Bicocca (UNIMIB) in Italy. Listed below, we will cover a number of phases of the work: (1) training a design to anticipate diagnosis from pathology images without defining the functions to utilize, so that it can discover what functions are essential; (2) penetrating that prognostic design utilizing explainability strategies; and (3) recognizing an unique function and confirming its association with client diagnosis. We explain this function and examine its usage by pathologists in our just recently released paper, “ Pathologist recognition of a machine-learned function for colon cancer danger stratification“. To our understanding, this is the very first presentation that medical professionals can discover brand-new prognostic functions from artificial intelligence, an appealing start for the future of this “knowing from deep knowing” paradigm.
Training a prognostic design to discover what functions are essential
One prospective technique to recognizing unique functions is to train ML designs to straight anticipate client results utilizing just the images and the paired result information. This remains in contrast to training designs to anticipate “intermediate” human-annotated labels for understood pathologic functions and after that utilizing those functions to anticipate results.
Preliminary work by our group revealed the expediency of training designs to straight anticipate diagnosis for a range of cancer types utilizing the openly offered TCGA dataset It was particularly amazing to see that for some cancer types, the design’s forecasts were prognostic after managing for offered pathologic and medical functions. Together with partners from the Medical University of Graz and the Biobank Graz, we consequently extended this work utilizing a big de-identified colorectal cancer accomplice. Translating these design forecasts ended up being an appealing next action, however typical interpretability strategies were challenging to use in this context and did not supply clear insights.
Translating the model-learned functions
To penetrate the functions utilized by the prognostic design, we utilized a 2nd design (trained to determine image resemblance) to cluster cropped spots of the big pathology images. We then utilized the prognostic design to calculate the typical ML-predicted danger rating for each cluster.
One cluster stood apart for its high typical danger rating (connected with bad diagnosis) and its unique visual look. Pathologists explained the images as including high grade growth (i.e., least-resembling typical tissue) in close distance to adipose (fat) tissue, leading us to call this cluster the “growth adipose function” (TAF); see next figure for comprehensive examples of this function. More analysis revealed that the relative amount of TAF was itself extremely and individually prognostic.
| Left: H&E pathology slide with an overlaid heatmap showing areas of the growth adipose function (TAF). Areas highlighted in red/orange are thought about to be most likely TAF by the image resemblance design, compared to areas highlighted in green/blue or areas not highlighted at all. Right: Agent collection of TAF spots throughout numerous cases. |
Confirming that the model-learned function can be utilized by pathologists
These research studies offered an engaging example of the capacity for ML designs to anticipate client results and a methodological technique for acquiring insights into design forecasts. Nevertheless, there stayed the interesting concerns of whether pathologists might discover and score the function recognized by the design while preserving verifiable prognostic worth.
In our newest paper, we worked together with pathologists from the UNIMIB to examine these concerns. Utilizing example pictures of TAF from the previous publication to discover and comprehend this function of interest, UNIMIB pathologists established scoring standards for TAF. If TAF was not seen, the case was scored as “missing”, and if TAF was observed, then “unifocal”, “multifocal”, and “extensive” classifications were utilized to show the relative amount. Our research study revealed that pathologists might reproducibly determine the ML-derived TAF which their scoring for TAF offered statistically substantial prognostic worth on an independent retrospective dataset. To our understanding, this is the very first presentation of pathologists discovering to determine and score a particular pathology function initially recognized by an ML-based technique.
Putting things in context: gaining from deep knowing as a paradigm
Our work is an example of individuals “gaining from deep knowing”. In conventional ML, designs gain from hand-engineered functions notified by existing domain understanding. More just recently, in the deep knowing age, a mix of massive design architectures, calculate, and datasets has actually allowed discovering straight from raw information, however this is typically at the expenditure of human interpretability. Our work couples using deep discovering to anticipate client results with interpretability techniques, to draw out brand-new understanding that might be used by pathologists. We see this procedure as a natural next action in the development of using ML to issues in medication and science, moving from using ML to boil down existing human understanding to individuals utilizing ML as a tool for understanding discovery.
Recognitions
This work would not have actually been possible without the efforts of coauthors Vincenzo L’Imperio, Markus Plass, Heimo Muller, Nicolò’ Tamini, Luca Gianotti, Nicola Zucchini, Robert Reihs, Greg S. Corrado, Dale R. Webster, Lily H. Peng, Po-Hsuan Cameron Chen, Marialuisa Lavitrano, David F. Steiner, Kurt Zatloukal, Fabio Pagni. We likewise value the assistance from Verily Life Sciences and the Google Health Pathology groups– in specific Timo Kohlberger, Yunnan Cai, Hongwu Wang, Kunal Nagpal, Craig Mermel, Trissia Brown, Isabelle Flament-Auvigne, and Angela Lin. We likewise value manuscript feedback from Akinori Mitani, Rory Sayres, and Michael Howell, and illustration assistance from Abi Jones. This work would likewise not have actually been possible without the assistance of Christian Guelly, Andreas Holzinger, Robert Reihs, Farah Nader, the Biobank Graz, the efforts of the slide digitization group at the Medical University Graz, the involvement of the pathologists who evaluated and annotated cases throughout design advancement, and the specialists of the UNIMIB group.